talk: Reverse Engineering of Dynamic Regulatory Networks from Morphological Data, 11am 4/7
Reverse Engineering of Dynamic Regulatory Networks
from Morphological Experimental Data
Prof. Daniel Lobo, Biological Sciences, UMBC
3:00pm 11:00am April 7 6th, ITE Building, Room 325b
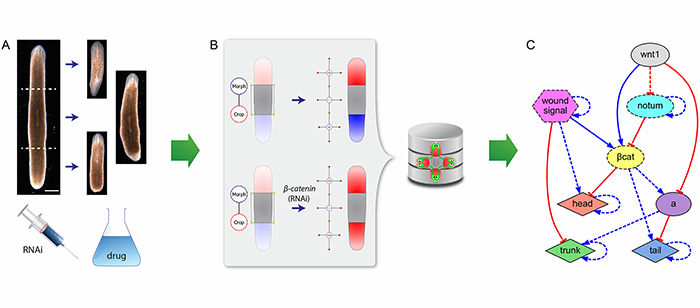
Many crucial experiments in developmental, regenerative, and cancer biology are based on manipulations and perturbations resulting in morphological outcomes. For example, planarian worms can regenerate a complete organism from almost any amputated piece, but knocking down certain genes can result in the regeneration of double-head phenotypes. However, the inherent complexity and non-linearity of biological regulatory networks prevent us from manually discerning testable comprehensive models from patterning and morphological results, and existent bioinformatics tools are generally limited to genomic or time-series concentration data. As a consequence, despite a huge experimental dataset in the literature, we still lack mechanistic explanations that can account for more than one or two morphological results in many model organisms. To bridge this gulf separating morphological data from an understanding of pattern and form regulation, we developed a computational methodology to automate the discovery of dynamic genetic networks directly from formalized phenotypic experimental data. In this seminar, I will present novel formal ontologies and databases of surgical, genetic, and pharmacological experiments with their resultant morphological phenotypes, together with artificial intelligence tools based on evolutionary computation and in silico simulators that can directly mine these data to reverse-engineer mechanistic dynamic genetic models. We demonstrated this approach by automatically discovering the first comprehensive model of planarian regeneration, which not only explains at once all the key experiments available in the literature (including surgical amputations, knock-down of specific genes, and pharmacological treatments), but also predicts testable novel pathways and genes. This approach is readily paving the way for understanding the regulation (and dis-regulation) of complex patterns and shapes in developmental, regenerative, and cancer biology.
Daniel Lobo is an Assistant Professor at the University of Maryland, Baltimore County. His research aims to understand, control, and design the dynamic regulatory mechanisms governing complex biological processes. To this end, his group develops new computational methods, ontologies, and high-performance in silico experiments to automate the reverse-engineering of quantitative models from biological data and the design of regulatory networks for specific functions. They seek to discover the mechanisms of development and regeneration, find therapies for cancer and other diseases, and streamline the application of synthetic biology. His work has received widespread media coverage including Wired, TechRadar, and Popular Mechanics.
Posted: March 29, 2016, 9:31 AM